Evaluating cell activation, killing and movement as a function of time
WORKFLOWS PLATFORM
Technology - Workflows
CellChorus provides comprehensive, dynamic analysis of single cells. Other methods of studying cellular activity lack the ability to integrate dynamic cellular behavior with molecular behavior at the single‑cell level. The company’s TIMING™ (Time-lapse Imaging Microscopy in Nanowell Grids) platform applies visual AI to evaluate cell activation, killing and movement as a function of time in order to maximize our understanding of cellular function, state and phenotype.
TIMING Workflow
After receiving cryopreserved effector and target cells from customers, CellChorus thaws and conducts cell line viability studies. CellChorus can also run customer‑specific assays to ensure that cells behave in an expected manner. At this point, CellChorus conducts its single‑cell TIMING workflow as follows:
1. Acquire images from nanowells
After cell line validation, cells are loaded into nanowell arrays, incubated and imaged with fluorescent microscopy.
2. Segment and track single cells
CellChorus uses visual AI to segment and track cells, and detect and quantify apoptosis and other cell-to-cell interactions.
3. Image secreted biomolecules
Bead-based sensors enable dynamic, time‑series imaging of secreted cytokines or other biomolecules.
4. Analyze across the TIMING dataset
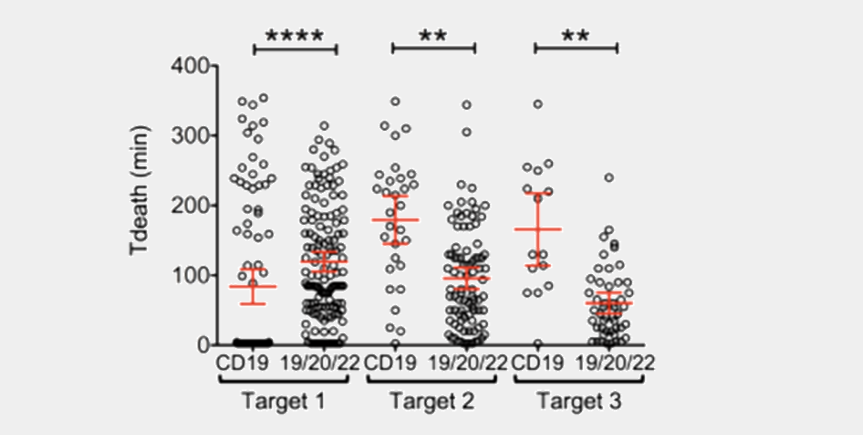
Analyzing the TIMING dataset across thousands of cells quantifies differences in cell behavior and interaction.
Multiomic Workflow
TIMING analysis enables CellChorus to identify cells of interest for further profiling—providing a more comprehensive picture of how your cells behave.
1. Perform TIMING assays
Perform time‑lapse imaging microscopy in nanowell grids (TIMING workflow).
2. Identify cells of interest
Identify and select cells of interest based on their dynamic behavior as a function of time.
3. Profile cells of interest
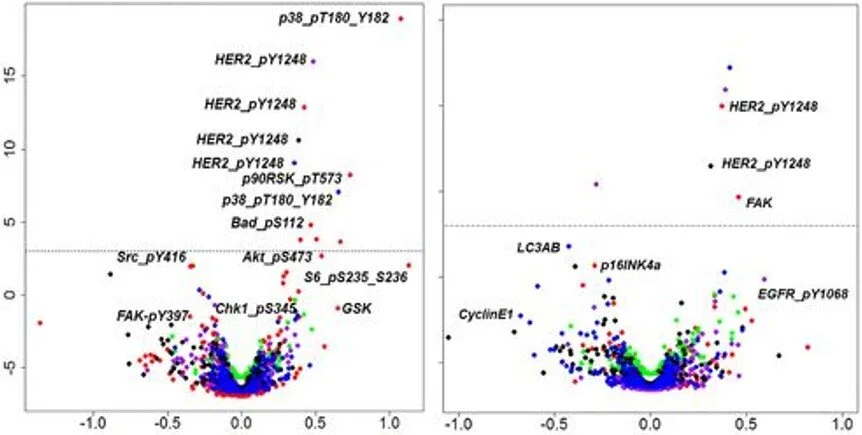
Perform molecular profiling such as single‑cell RNA sequencing on cells of interest.
4. Integrate data
Integrate TIMING data of dynamic cellular behavior with single‑cell molecular profiling data.
See an example of the CellChorus workflow from Cancer Immunology Research.
The CellChorus platform is broadly applicable across a variety of cell types and applications. Whereas life science researchers have benefitted from significant advances in single-cell analysis, CellChorus goes beyond static assaying. With TIMING, life sciences innovators can watch new biology and gain new and unexpected understandings of how cells move, activate, kill and survive.
The company’s technology is protected in part by issued patents and pending patent applications, including US/10,746,736.